-
Notifications
You must be signed in to change notification settings - Fork 0
/
Copy pathextinction.Rmd
472 lines (361 loc) · 11.7 KB
/
extinction.Rmd
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
302
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
346
347
348
349
350
351
352
353
354
355
356
357
358
359
360
361
362
363
364
365
366
367
368
369
370
371
372
373
374
375
376
377
378
379
380
381
382
383
384
385
386
387
388
389
390
391
392
393
394
395
396
397
398
399
400
401
402
403
404
405
406
407
408
409
410
411
412
413
414
415
416
417
418
419
420
421
422
423
424
425
426
427
428
429
430
431
432
433
434
435
436
437
438
439
440
441
442
443
444
445
446
447
448
449
450
451
452
453
454
455
456
457
458
459
460
461
462
463
464
465
466
467
468
469
470
471
---
title: '11.18'
output: html_document
---
```{r setup, include=FALSE}
knitr::opts_chunk$set(echo = TRUE)
```
## R Markdown
This is an R Markdown document. Markdown is a simple formatting syntax for authoring HTML, PDF, and MS Word documents. For more details on using R Markdown see <http://rmarkdown.rstudio.com>.
When you click the **Knit** button a document will be generated that includes both content as well as the output of any embedded R code chunks within the document. You can embed an R code chunk like this:
```{r cars}
summary(cars)
```
## Including Plots
You can also embed plots, for example:
```{r pressure, echo=FALSE}
plot(pressure)
```
Note that the `echo = FALSE` parameter was added to the code chunk to prevent printing of the R code that generated the plot.
genna's extinction code prior to 11.18"
---
title: "Extinctions Unit"
author: "Genna Fudin, Athena Sabaria, Luanne Yang"
maketitle: true
output: github_document
---
```{r include=FALSE}
library("tidyverse")
library("httr")
library("jsonlite")
#library("printr")
knitr::opts_chunk$set(comment=NA)
```
```{r setup}
library("tidyverse")
library("httr")
library("jsonlite")
knitr::opts_chunk$set(comment=NA)
```
```{r}
resp <- GET("https://apiv3.iucnredlist.org/api/v3/speciescount?token=9bb4facb6d23f48efbf424bb05c0c1ef1cf6f468393bc745d42179ac4aca5fee")
```
```{r}
base_url <- "https://apiv3.iucnredlist.org"
endpoint <- "/api/v3/species/page/"
page_number <- 0
args <- "?token="
token <- "9bb4facb6d23f48efbf424bb05c0c1ef1cf6f468393bc745d42179ac4aca5fee"
query <- paste0(base_url, endpoint, page_number, args, token)
```
```{r}
resp <- GET(query)
```
```{r}
txt <- content(resp,as = "parsed")
```
```{r}
# x <- txt$result[[1]]
# data.frame(sci_name = x$scientific_name)
all_sci_names <-
purrr::map_dfr(txt$result,
function(x)
data.frame(sci_name = x$scientific_name,
kingdom = x$kingdom_name)
)
all_sci_names %>% count(kingdom)
```
```{r}
resp <- map(query, GET)
```
```{r}
status <- map_int(resp, status_code)
good <- resp[status==200]
try_again <- map(query[status != 200], GET)
good <- c(good, try_again)
```
```{r}
txts <- map(good, content, as = "parsed")
```
```{r}
rectangle_me <- function(x) as_tibble(compact(x))
stack_txt <- function(txt) map_dfr(txt$result, rectangle_me)
all_sci_names <- map_dfr(txts, stack_txt)
all_sci_names
```
```{r}
extinct <- all_sci_names %>% filter(category == "EX") %>% arrange(scientific_name)
extinct
```
```{r}
name <- extinct$scientific_name[[10]]
url <- paste0(base_url, "/api/v3/species/narrative/", name, args, token)
x <- url %>% GET() %>% content()
rationale <- x$result[[1]]$rationale
library(stringr)
stringr::str_extract(rationale, "\\d+")
```
## Extinctions Module
_Are we experiencing the sixth great extinction?_
What is the current pace of extinction? Is it accelerating? How does it compare to background extinction rates?
## Background
- [Section Intro Video](https://youtu.be/QsH6ytm89GI)
- [Ceballos et al (2015)](http://doi.org/10.1126/sciadv.1400253)
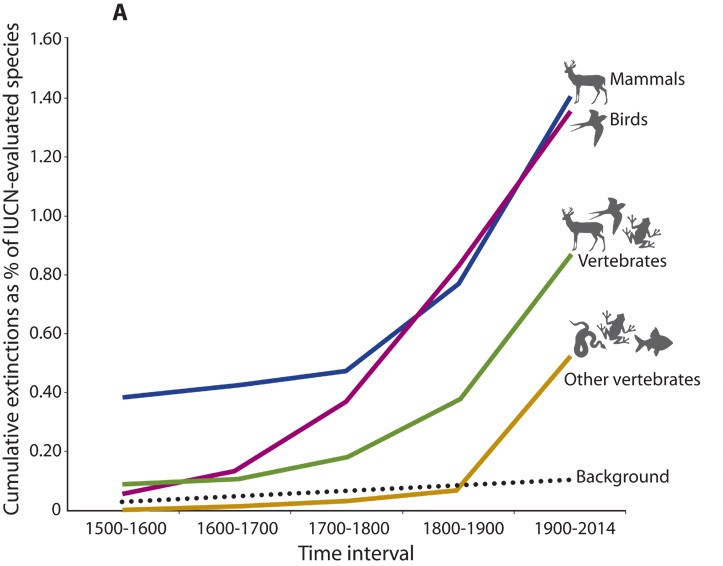
Our focal task will be to reproduce the result from Ceballos and colleagues showing the recent increase in extinction rates relative to the background rate:
## Computational Topics
- Accessing data from a RESTful API
- Error handling
- JSON data format
- Regular expressions
- Working with missing values
## Additional references:
- http://www.hhmi.org/biointeractive/biodiversity-age-humans (Video)
- [Barnosky et al. (2011)](http://doi.org/10.1038/nature09678)
- [Pimm et al (2014)](http://doi.org/10.1126/science.1246752)
- [Sandom et al (2014)](http://dx.doi.org/10.1098/rspb.2013.3254)
```{r}
base_url <- "https://apiv3.iucnredlist.org"
endpoint <- "/api/v3/species/page/"
page_number <- 0:12
args <- "?token="
token <- "9bb4facb6d23f48efbf424bb05c0c1ef1cf6f468393bc745d42179ac4aca5fee"
query <- paste0(base_url, endpoint, page_number, args, token)
```
```{r}
content(resp,as = "parsed")
```
```{r}
?content
```
It "parses" the JSON data structure returned by the API into an R list objec
Prof Boettiger 7 days ago
recall that the first endpoint, https://apiv3.iucnredlist.org/api/v3/docs#species-count , tells you how many records total
Prof Boettiger 7 days ago
and the species endpoint returns them in 10,000 species per chunk.
Prof Boettiger 7 days ago
If you wanted to be really fancy, you could use the result of the first call to automatically determine how many pages you'd need to iterate through. The nice part about that is your code would still work even when enough species are added to the database to need an additional page.
Prof Boettiger 7 days ago
https://apiv3.iucnredlist.org/api/v3/speciescount?token=9bb4facb6d23f48efbf424bb05c0c1ef1cf6f468393bc745d42179ac4aca5fee
Alexlopitz 7 days ago
So yeah for anyone else reading this 122933 species which would indicate 13 total pages from indexes 0:12
```{r}
txt <- map(many_resp, content, as = "parsed")
```
No encoding supplied: defaulting to UTF-8.
base_url <- "https://apiv3.iucnredlist.org"
endpoint <- "/api/v3/species/page/"
page_number <- 0:12
args <- "?token="
token <- "9bb4facb6d23f48efbf424bb05c0c1ef1cf6f468393bc745d42179ac4aca5fee"
## paste0 is vectorized:
queries <- paste0(base_url, endpoint, page_number, args, token)
```{r}
for (i in 1:13){
resp <- paste("resp_pg", i-1, sep = "")
assign(resp, GET(query[i]))
}
```
```{r}
queries <- map(query, GET)
```
```{r}
resps <- map(queries, GET)
```
```{r}
map_int(resps, status_code)
```
```{r}
create_df <- function(resp) {
txt <- content(resp, as = "parsed")
all_sci_names <-
purrr::map_dfr(txt$result,
function(x)
data.frame(sci_name = x$scientific_name,
kingdom = x$kingdom_name)
)
return(all_sci_names)
}
# lis <- c()
# for(i in 0:12) {
# resp <- paste("resp_pg", i, sep = "")
# lis <- c(lis, resp)
# }
lis <- c(resp_pg0, resp_pg1, resp_pg2, resp_pg3, resp_pg4, resp_pg5, resp_pg6, resp_pg7, resp_pg8, resp_pg9, resp_pg10, resp_pg11, resp_pg12)
```
blah blah blahh
Code from before 11.30 before I pulled Luanne's update from GitHub
---
title: "Extinctions Unit"
author: "Genna Fudin, Athena Sabaria, Luanne Yang"
maketitle: true
output: github_document
---
```{r include=FALSE}
library("tidyverse")
library("httr")
library("jsonlite")
library("stringr")
#library("printr")
knitr::opts_chunk$set(comment=NA)
```
## Extinctions Module
_Are we experiencing the sixth great extinction?_
What is the current pace of extinction? Is it accelerating? How does it compare to background extinction rates?
## Background
- [Section Intro Video](https://youtu.be/QsH6ytm89GI)
- [Ceballos et al (2015)](http://doi.org/10.1126/sciadv.1400253)
Our focal task will be to reproduce the result from Ceballos and colleagues showing the recent increase in extinction rates relative to the background rate:

## Computational Topics
- Accessing data from a RESTful API
- Error handling
- JSON data format
- Regular expressions
- Working with missing values
## Additional references:
- http://www.hhmi.org/biointeractive/biodiversity-age-humans (Video)
- [Barnosky et al. (2011)](http://doi.org/10.1038/nature09678)
- [Pimm et al (2014)](http://doi.org/10.1126/science.1246752)
- [Sandom et al (2014)](http://dx.doi.org/10.1098/rspb.2013.3254)
```{r}
base_url <- "https://apiv3.iucnredlist.org"
endpoint <- "/api/v3/species/page/"
page_number <- 0:12
args <- "?token="
token <- "9bb4facb6d23f48efbf424bb05c0c1ef1cf6f468393bc745d42179ac4aca5fee"
query <- paste0(base_url, endpoint, page_number, args, token)
```
```{r}
for (i in 1:13){
resp <- paste("resp_pg", i-1, sep = "")
assign(resp, GET(query[i]))
}
```
```{r}
m <- map(query, GET)
```
```{r}
create_df <- function(resp) {
txt <- content(resp, as = "parsed")
all_sci_names <-
purrr::map_dfr(txt$result,
function(x)
data.frame(sci_name = x$scientific_name,
kingdom = x$kingdom_name)
)
return(all_sci_names)
}
# lis <- c()
# for(i in 0:12) {
# resp <- paste("resp_pg", i, sep = "")
# lis <- c(lis, resp)
# }
lis <- c(resp_pg0, resp_pg1, resp_pg2, resp_pg3, resp_pg4, resp_pg5, resp_pg6, resp_pg7, resp_pg8, resp_pg9, resp_pg10, resp_pg11, resp_pg12)
```
```{r}
#livecode all below
resp <- map(query, GET)
```
```{r}
status <- map_int(resp, status_code)
good <- resp[status==200]
try_again <- map(query[status != 200], GET)
good <- c(good, try_again)
```
```{r}
txts <- map(good, content, as = "parsed")
```
```{r}
rectangle_me <- function(x) as_tibble(compact(x))
stack_txt <- function(txt) map_dfr(txt$result, rectangle_me)
all_sci_names <- map_dfr(txts, stack_txt)
all_sci_names
```
```{r}
##from Luanne/Athena's code
##birds <- all_sci_names %>% select(class_name == "AVES")
##vert <- all_sci_names %>% select(class_name != "AVES")
```
extinct_m <- all_sci_names %>% filter(category == "EX", class_name == 'MAMMALIA') %>% arrange(scientific_name)
name_m <- extinct_m$scientific_name
url <- paste0(base_url, "/api/v3/species/narrative/", name_m, args, token)
resp2 <- map(url, GET)
narrative <- map(resp2, content, as = "parsed")
get_rationale <- function(x) x$result[[1]]$rationale
safe_get <- safely(get_rationale, otherwise = "")
rationale_txt <- map(narrative, safe_get)
```
years <- stringr::str_match_all(rationale_txt, "(\\d{2})\\d{2}")
all(years[1][[1]][,2] == '19')
```{r}
extinct <- all_sci_names %>% filter(category == "EX") %>% arrange(scientific_name)
name <- extinct$scientific_name[1:919]
url <- paste0(base_url, "/api/v3/species/narrative/", name, args, token)
resp2 <- map(url, GET)
narrative <- map(resp2, content, as = "parsed")
get_rationale <- function(x) x$result[[1]]$rationale
safe_get <- safely(get_rationale, otherwise = "")
rationale_txt <- map(narrative, safe_get)
#rationale_txt <- map(narrative, get_rationale)
```
```{r}
# rando <- c()
# for (i in 1:13){
# name <- extinct$scientific_name[[i]]
# url <- paste0(base_url, "/api/v3/species/narrative/", name, args, token)
# x <- url %>% GET() %>% content()
# rationale <- x$result[[1]]$rationale
# rando <- c(rando, rationale)
# }
#
# stringr::str_extract(rando, "\\d+")
```
From 11.18
Carl's Live Code
```{r}
download.file("https://github.com/espm-157/extinction-template/releases/download/data/resp2.rds", "resp2.rds")
```
```{r}
resp2 <- readRDS("resp2.rds")
```
```{r}
status <- map_int(resp2, status_code)
all(status == 200)
narrative <- map(resp2, content)
names <- map(narrative, "name")
missing <- map_lgl(names, is.null)
good_names <- names[!missing]
good_narrative <- narrative[!missing]
result <- map(good_narrative, "result")
result1 <- map(result, function(x) x[[1]])
rationale <- map(result1, "rationale")
missing_rationale <- map_lgl(rationale, is.null)
complete_narrative <- good_narrative[!missing_rationale]
complete_rationale <- rationale[!missing_rationale]
complete_names <- good_names[!missing_rationale]
narrative_df <- tibble(scientific_name = as.character(complete_names),
rationale = as.character(complete_rationale))
sum(missing_rationale)
```
```{r}
narrative_final <- narrative_df %>%
left_join(extinct) %>%
mutate(date = stringr::str_extract(rationale, "\\d{4}"),
century = stringr::str_extract(date, "\\d{2}"))
```
## Creating a new column for the y-axis on graph (cumulative value)
```{r}
year_bin <- c("1500-1600", "1600-1700", "1700-1800", "1800-1900", "1900-2014")
```
Change column date into integer columm
Cut function?
```{r}
narrative_final$date <- as.numeric(as.character(narrative_final$date))
```
as_int
Mammals, birds, vertebrates, other vertebrates
## Add 1 to centuries
```{r}
ggplot(data = narrative_df, aes(x = "Century", y = ))
```