| marp | theme | class | paginate | backgroundColor | color | header | footer |
|---|---|---|---|---|---|---|---|
true |
default |
lead |
true |
Scientific AI and the Future of OME-Zarr |
[](https://fideus.io) Matt McCormick, PhD | fideus labs | EMBL BIA 2025 |
Matt McCormick, PhD fideus labs
EMBL Advanced Methods in Bioimage Analysis September 17, 2025
🌐 HTML slides | 📄 PDF slides | 📂 GitHub repository
50 minutes + 10 minutes Q&A
-
Extended introduction to ngff-zarr (15 min)
- Converting bioimages to OME-Zarr
-
Introduction to MCP Servers (15 min)
- Add the ngff-zarr MCP server to agentic AI tools
-
The ngff-zarr MCP Server in Action (15 min)
- AI-powered conversions and batch processing
-
fideus labs introduction (5 min)
Next-Generation Scientific Imaging
What is OME-Zarr?
- Cloud-native bioimaging file format from the Open Microscopy Environment (OME)
- Built on Zarr - chunked, compressed array storage
- Multiscale pyramidal data structure
- Interoperable across platforms and tools
- FAIR data principles: Findable, Accessible, Interoperable, Reusable
Traditional Problems:
- 🏭 Vendor-specific proprietary formats
- 📦 Monolithic files difficult to stream
- ☁️ Limited cloud compatibility
- 🐢 Poor scalability for large datasets
OME-Zarr Solutions:
- 📖 Open specification
- 🧩 Chunked data access
- 🌐 Cloud-optimized storage
- ⚡ Parallel processing friendly
- ngff-zarr is an lean and kind open-source toolkit for working with OME-Zarr, the next-generation file format for scientific imaging.
- Provides command-line, Python, TypeScript, and AI interfaces for converting, validating, optimizing, and analyzing bioimaging data.
- Developed by the OME-Zarr and ITK communities for interoperability and performance.
- Supports a wide range of scientific image formats and workflows.
- 🔄 Convert your scientific images (NRRD, TIFF, HDF5, and more) to OME-Zarr for scalable, cloud-ready storage.
- ✅ Validate OME-Zarr datasets to ensure compliance and interoperability.
- 🛠️ Optimize chunking and compression for efficient access and storage.
- 🤖 Integrate with AI and analysis tools via the Model Context Protocol (MCP).
- 🚀 Automate batch processing and reproducible workflows for large-scale projects.
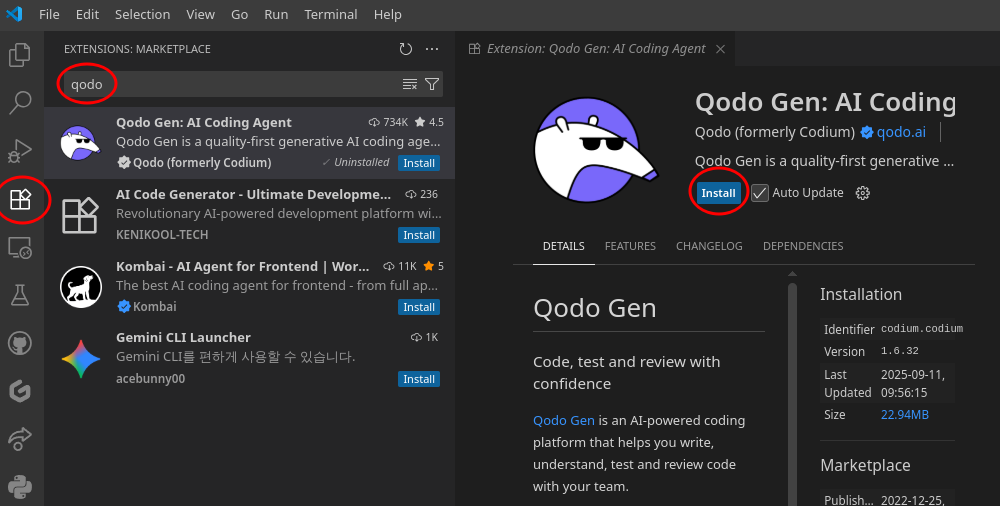
Install Visual Studio Code
Download VS Code:
- 🌐 Web: Visit code.visualstudio.com
- 🐧 Linux:
sudo snap install code --classicor download .deb/.rpm - 🍎 macOS: Download from website or
brew install --cask visual-studio-code - 🪟 Windows: Download installer or
winget install Microsoft.VisualStudioCode
Pixi is a fast, modern, and reproducible package and environment manager built on the conda ecosystem. It provides:
- 🚀 Easy, reproducible environments for any language
- 🛠️ Task runner for project automation
- 🔒 Isolation and cross-platform support (Linux, macOS, Windows)
- 📦 Simple dependency management with a single file (
pixi.tomlorpyproject.toml)
On Linux/macOS:
wget -qO- https://pixi.sh/install.sh | shOn Windows (PowerShell):
powershell -ExecutionPolicy ByPass -c "irm -useb https://pixi.sh/install.ps1 | iex"After installation, add ~/.pixi/bin (Linux/macOS) or %USERPROFILE%\.pixi\bin (Windows) to your PATH if not done automatically.
Pixi lets you define and run project tasks in your pixi.toml or pyproject.toml.
To run a task (e.g., start):
pixi run startYou can define custom tasks (like test, lint, etc.) and run them the same way:
pixi run test
pixi run lintPixi ensures all dependencies and the environment are set up before running your task.
Enter an interactive shell with your project environment activated:
pixi shellWhat happens:
- 🔧 Environment activated - all dependencies available
- 🎯 Direct command execution - no need for
pixi runprefix - 🚪 Easy exit - just type
exitwhen done
pixi run convert- 🔍 Automatic multiscale generation - without aliasing artifacts
- 🧩 Intelligent chunking - optimized for access patterns
- 📊 Metadata preservation - spatial information maintained
- 🗜️ Compression applied - reduced file size
- ☁️ Cloud-ready format - object-store optimized, can be served via HTTP
pixi run convert-ome-zarr-0.5# Count the number of files created
find carp.ome.zarr -type f | wc -lpixi run convert-sharding# Count the number of files created
find carp.ome.zarr -type f | wc -l- 🪣 Sharding enabled - multiple chunks stored in single files
- 📦 Optimized storage - fewer small files, better filesystem performance
What is Sharding? Sharding combines multiple small chunks into larger "shard" files, dramatically reducing the number files needed to store data while maintaining random access capabilities.
Connecting AI to Your Data
What is Model Context?
- 📝 Information the AI model can "see" and reason about
- 🧮 Limited capacity - typically measured in tokens (words/symbols)
- ⏱️ Temporary memory - context is conversation-specific
- 🎯 Scope of knowledge for making informed decisions
Why Context Matters:
- 🔍 Better understanding - more relevant, accurate responses
- 🎛️ Tool selection - AI chooses appropriate tools for the task
- 🔗 Data integration - combines multiple information sources
- 🚀 Workflow automation - maintains state across complex operations
The Challenge: How do we give AI access to your scientific data and tools?
Universal standard for connecting AI assistants to external data and tools
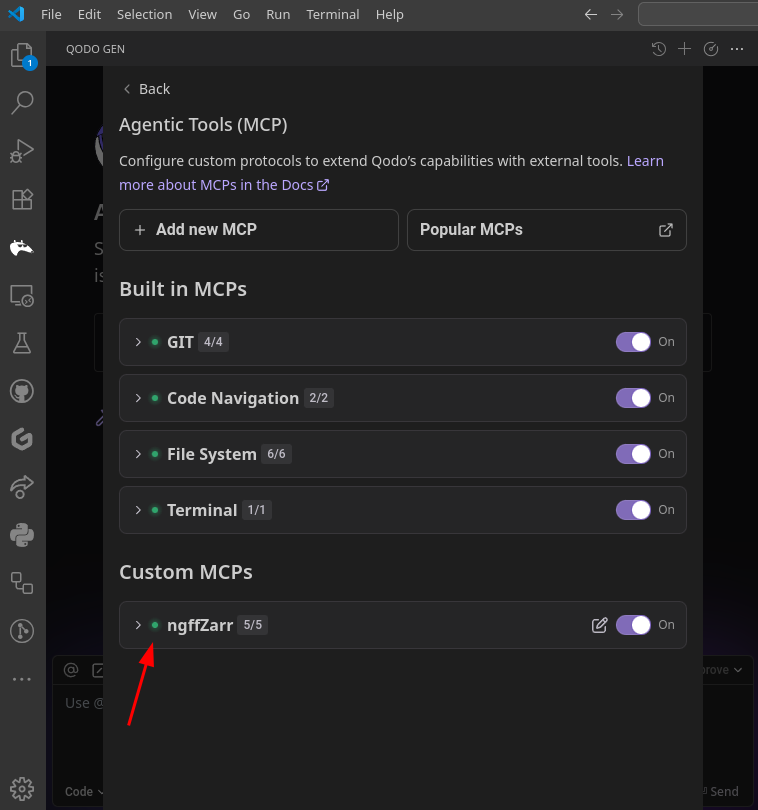
Key Components:
- 🤖 MCP Client - integrated in AI applications
- 🖥️ MCP Server - exposes specific capabilities
- 🔗 Transport Layer - JSON-RPC 2.0 communication
- 🔧 Standardized Interface - tools, resources, prompts
AI Application (Qodo, Claude, etc.)
↕️ JSON-RPC 2.0
MCP Client
↕️ STDIO/HTTP
MCP Server (ngff-zarr)
↕️
Scientific Data & Tools
Benefits:
- Single protocol for all integrations
- Bidirectional communication
- Context-aware AI interactions
Before MCP:
- 🔧 Custom integrations for each tool
- 🚫 Limited AI access to scientific data
- ✋ Manual, error-prone workflows
With MCP:
- 💬 Natural language interface to scientific tools
- 🤖 Automated data processing pipelines
- 🧠 AI-driven optimization and analysis
- 🔄 Reproducible computational workflows
Install uv, if not already installed
pixi global install uvuvx, which comes with uv, will be used to install the ngff-zarr-mcp command-line tool and its dependencies, and run the MCP server.
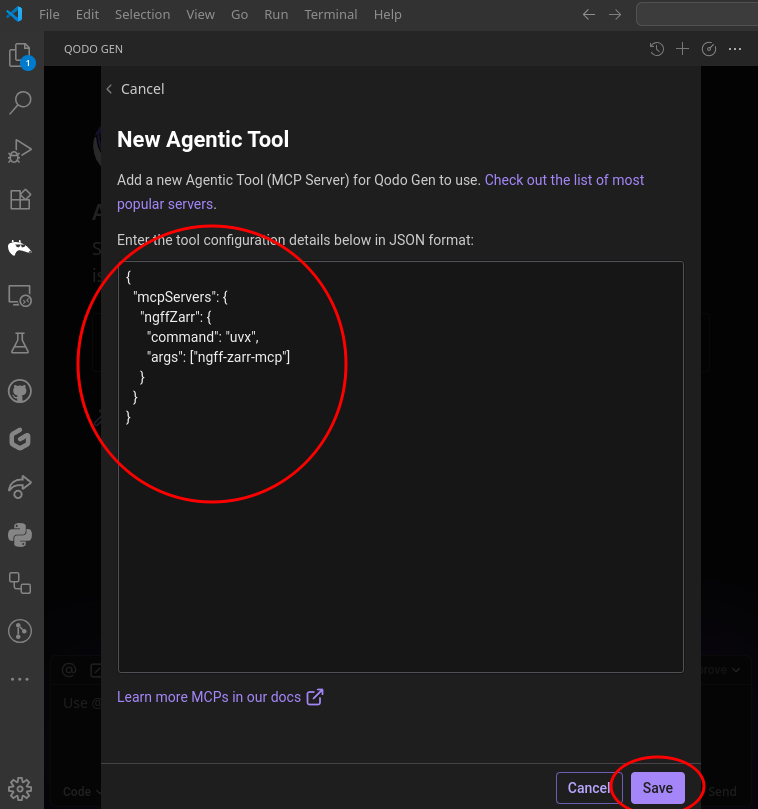
{
"mcpServers": {
"ngffZarr": {
"command": "uvx",
"args": ["ngff-zarr-mcp"]
}
}
}AI-Powered Scientific Image Processing
Core Functions:
- 🔄 Convert scientific formats to OME-Zarr
- 🔍 Inspect and validate OME-Zarr stores
- 🛠️ Optimize compression and chunking
- 📝 Generate processing scripts
- 📦 Batch operation planning
AI Integration:
- 💬 Natural language commands
- 🎯 Intelligent parameter selection
- 🤖 Automated workflow generation
Put the Qodo Anteater to work!
In Qodo chat:
Convert the vs_male.nrrd file to OME-Zarr format and
find the optimal compression codec for this type of data.
- 🔍 Analyze the input file
- 🎯 Select appropriate parameters
- ⚙️ Execute the conversion
- 📊 Report optimization results
Ask the AI to explore:
Examine the contents of carp.ome.zarr and tell me
about its structure, dimensions, and metadata
- 🔍 Inspect multiscale levels
- 📏 Report spatial metadata
- 🧩 Analyze chunk structure
- ✨ Suggest next steps
Scale up with AI automation:
I have a folder of 50 similar NRRD files.
Generate a Python script to batch convert them all
to OME-Zarr with the same optimal settings
- 🐍 Complete Python script
⚠️ Error handling- 📈 Progress reporting
- 🎯 Optimized parameters from previous analysis
Today's Demo Shows:
- 💬 Conversational scientific computing
- 🤖 Automated optimization
- 🔄 Reproducible workflows
- ✨ Accessible advanced techniques
Tomorrow's Possibilities:
- 🧬 Multi-modal analysis pipelines
- 🧠 Intelligent experiment design
- 🛡️ Automated quality control
- 🌐 Cross-platform integration
Fostering trust 🤝 and advancing understanding 🧠 from scientific and biomedical images 🔬️
About fideus labs
Specialties:
- Biomedical Imaging - ITK core development
- Scientific Visualization - advanced rendering
- Open science - pioneering decentralized science
- AI Integration - intelligent workflows
Open Source Leadership:
- ITK (Insight Toolkit) core team
- OME-Zarr ecosystem contributor
- Curate ngff-zarr development
Research Partnership:
- Government laboratories
- Academic institutions
- Industry leaders
- Open source communities
fideus labs services:
- Custom imaging solutions
- Scientific software development
- Training and consultation
Connect
We are hiring! Send us your CV and GitHub profile.
✅ OME-Zarr - Future of scientific imaging formats
✅ MCP Servers - Bridge AI and scientific tools
✅ Natural Language - New interface for scientific computing
✅ Accessible Research - Cloud-native, collaborative science
What we covered:
- OME-Zarr fundamentals and conversion
- MCP architecture and benefits
- AI-powered scientific workflows
Let's discuss:
- Your specific use cases
- Integration challenges
- Future possibilities
- Next steps for implementation