This library performs unsupervised batch correction of high-dimensional data via the use of mutual nearest neighbors (MNNs).
MNN correction was initially described in the context of single-cell RNA sequencing data analysis (see Haghverdi et al., 2018)
but the same methodology can be applied for any high-dimensional data containing shared populations across multiple batches.
The MNN implementation here is based on the fastMNN() function in the batchelor package,
which provides a number of improvements and speed-ups over the original method in the Haghverdi paper.
Consider a dense matrix in column-major format where rows are dimensions (e.g., principal components) and cells are columns,
and a vector of integers specifying the batch of origin for each cell.
These are supplied to the mnncorrect::compute() function to compute corrected values:
#include "mnncorrect/MnnCorrect.hpp"
std::vector<double> matrix(ndim * nobs); // fill with values...
std::vector<int> batch(nobs) // fill with batch IDs from [0, num_batches)
mnncorrect::Options<int, double> opt;
std::vector<double> output(ndim * nobs);
mnncorrect::compute(ndim, nobs, matrix.data(), batch.data(), output.data(), opt);We also support batches in separate arrays, storing the corrected values for all batches in a single output array:
int nbatches = 3;
std::vector<int> batch_size;
std::vector<std::vector<double> > batches;
for (int b = 0; b < 3; ++b) { // mocking up three batches of different size.
batch_size.push_back((b + 1) * 100);
batch.resize(ndim * batch_size.back()); // fill with values...
}
std::size_t total_size = std::accumulate(batch_size.begin(), batch_size.end(), 0);
std::vector<double> output(ndim * total_size);
mnncorrect::compute(ndim, batch_size, batch_ptrs, output.data(), opt);Advanced users can also fiddle with the options:
// Number of neighbors to use for MNN identification.
opt.num_neighbors = 10;
// Parallelization of various calculations, e.g., neighbor search.
opt.num_threads = 3;
// Number of recursive steps for calculation of the center of mass.
opt.num_steps = 2;
// Change the nearest-neighbor search algorithm:
opt.builder.reset(new knncolle_annoy::AnnoyBuilder<Annoy::Euclidean>);See the reference documentation for more details.
We assume that (i) our batches share some subpopulations, and (ii) even after the addition of an arbitrary batch effect, the cells from one subpopulation in one batch are still closer to the cells in the corresponding subpopulation in the other batch (and vice versa) when compared to cells in different subpopulations. Thus, by identifying pairs of cells that are MNNs, we can determine which subpopulations are shared across batches. Any differences in location between batches for the shared subpopulations are attributed to batch effects and targeted for removal. In contrast, a subpopulation unique to a single batch will not contain any MNNs (and thus will not interfere with correction), as it will not have a corresponding subpopulation in the other batch for which it can be the closest neighbor.
To remove batch effects, we consider one batch to be the "reference" and another to be the "target".
For each MNN pair, we compute a correction vector that moves the target batch towards the reference.
For each cell
The correction vector for each MNN pair is not directly computed from its two paired cells.
Rather, for each cell, we compute a "center of mass" using neighboring points from the same batch.
Most simply, the center of mass is defined as the mean coordinates of the
In the case of >2 batches, we define a merge order based on the batch size, variance, residual sum of squares, or the input order. For the first batch to be merged, we identify MNN pairs to all other batches at once. The subsequent correction effectively distributes the first batch's cells to all other batches. This process is repeated for all remaining batches until only one batch remains that contains all cells. By using all batches to identify MNN pairs at each step, we improve the chance of correctly matching subpopulations across batches, even if they are missing from certain batches.
The tests/R/examples directory contains a few examples using the C++ code on some real datasets (namely, single-cell RNA-seq datasets).
To run these, install the package at tests/R/package (this requires the scran.chan package, which also wraps this C++ library in a more complete package).
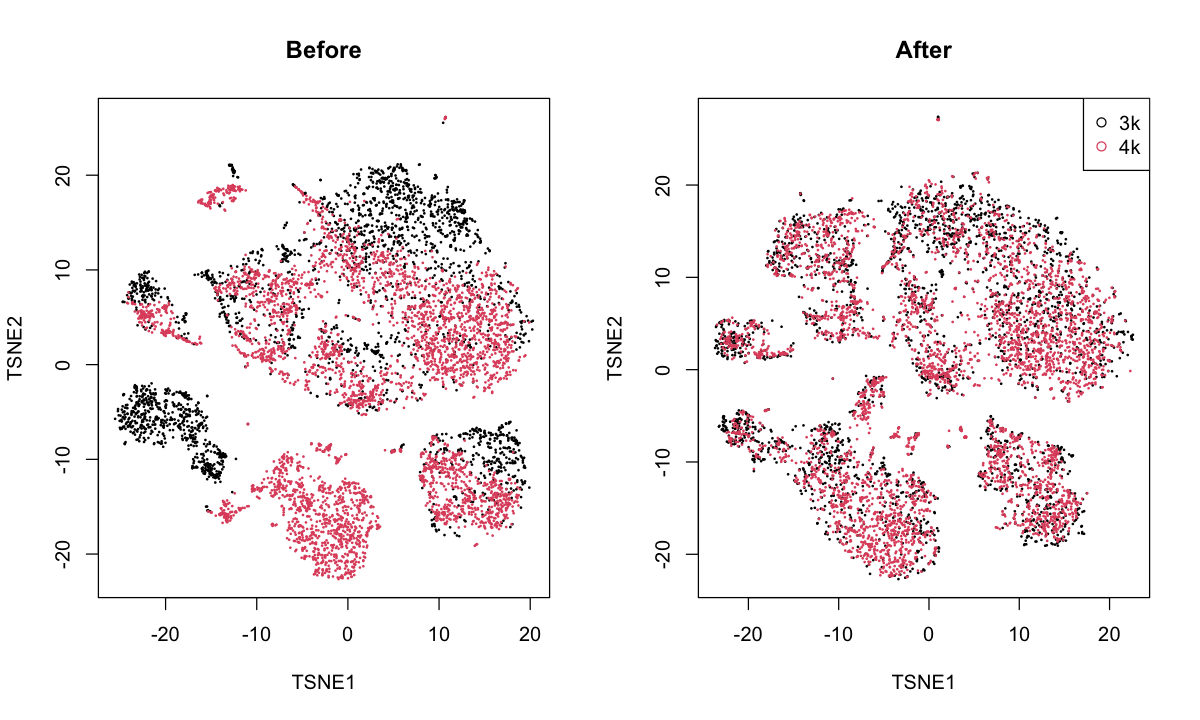
pbmc: mergesthe PBMC 3K and 4K datasets from 10X Genomics.
These are technical replicates (I think) so a complete merge is to be expected.
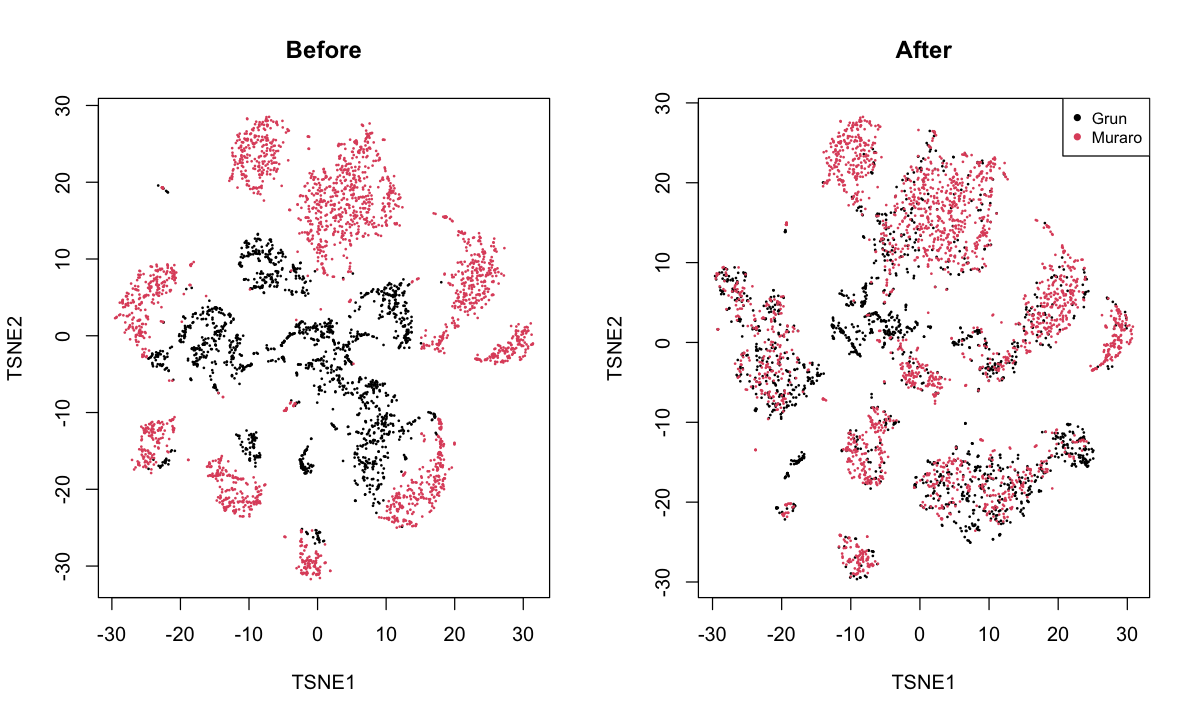
pancreas: merges the Grun et al. (2016) and Muraro et al. (2016) datasets.
I believe this involves data from different patients but using the same-ish technology.
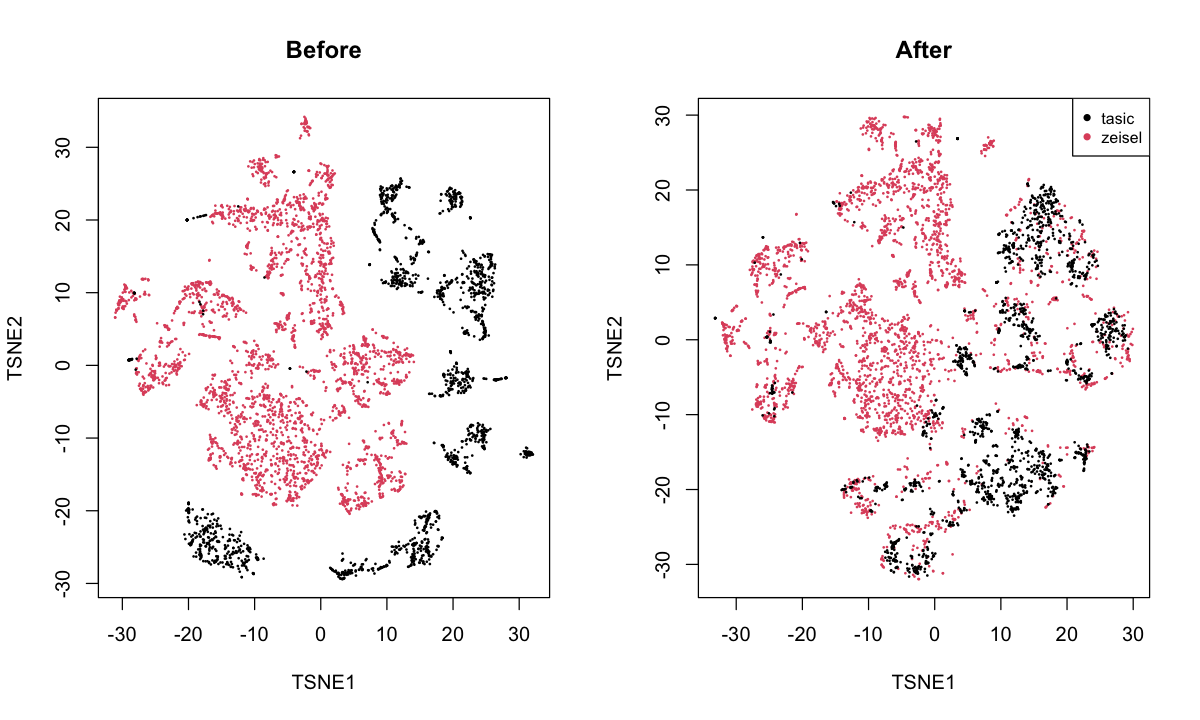
neurons: merges the Zeisel et al. (2015) and Tasic et al. (2016) datasets.
This involves different technologies and different cell populations.
If you're using CMake, you just need to add something like this to your CMakeLists.txt:
include(FetchContent)
FetchContent_Declare(
mnncorrect
GIT_REPOSITORY https://github.com/libscran/mnncorrect
GIT_TAG master # replace with a pinned release
)
FetchContent_MakeAvailable(mnncorrect)Then you can link to libscran to make the headers available during compilation:
# For executables:
target_link_libraries(myexe mnncorrect)
# For libaries
target_link_libraries(mylib INTERFACE mnncorrect)By default, this will use FetchContent to fetch all external dependencies.
Applications should consider pinning versions of dependencies for stability - see extern/CMakeLists.txt for suggested versions.
If you want to install them manually, use -DMNNCORRECT_FETCH_EXTERN=OFF.
find_package(libscran_mnncorrect CONFIG REQUIRED)
target_link_libraries(mylib INTERFACE libscran::mnncorrect)To install the library, use:
mkdir build && cd build
cmake .. -DMNNCORRECT_TESTS=OFF
cmake --build . --target installAgain, this will use FetchContent to retrieve dependencies, see comments above.
If you're not using CMake, the simple approach is to just copy the files - either directly or with Git submodules - and include their path during compilation with, e.g., GCC's -I.
This also requires the external dependencies listed in extern/CMakeLists.txt.
Haghverdi L, Lun ATL, Morgan MD, Marioni JC (2018). Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 36(5):421-427